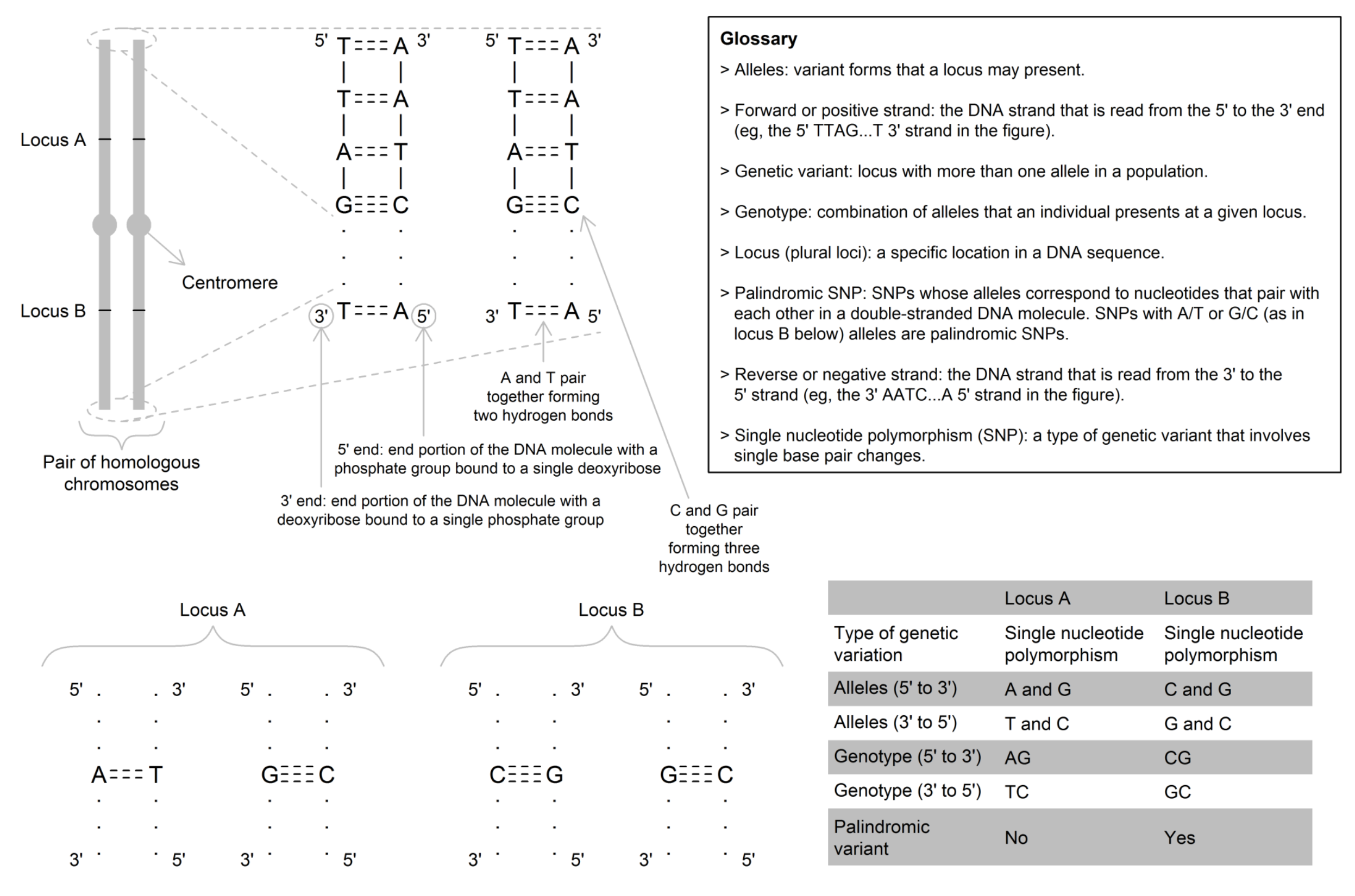
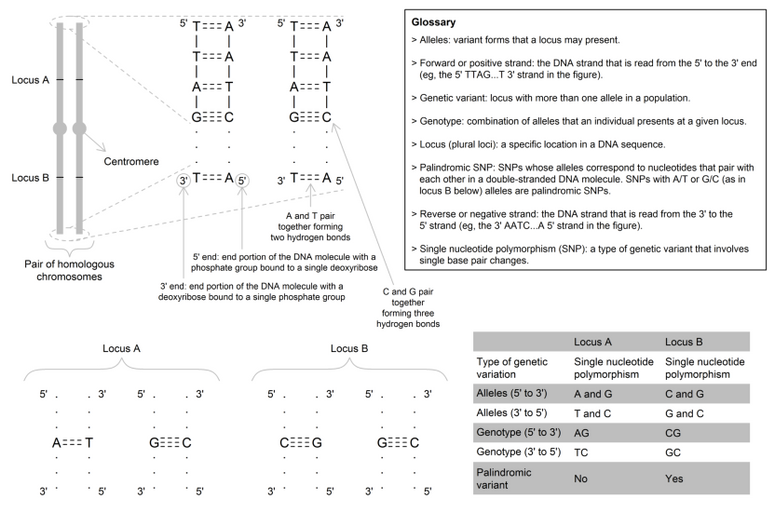
A variant form of a single nucleotide polymorphism (SNP), a specific polymorphic site or a whole gene detectable at a locus. Most SNPs are biallelic, meaning that there are two values (forms) that are possible (e.g., the rs234 SNP denotes that individuals could have either an A or G allele at position 105,920,689 on chromosome 7 on the forward strand). At a given SNP, the most common allele is referred to as the "major allele"; whereas, the least common (or rarer) allele is referred to as the "minor allele". The minor allele frequency is therefore the frequency at which the minor allele occurs within a population.
In MR, most commonly, the genetic instrumental variable (IV) is one or more SNP(s) that are robustly associated with the exposure of interest. The number of "trait-increasing" alleles is used analogously to the lifetime "dose" of the exposure. The commonest unit of the genetic IV-exposure or genetic IV-outcome associations are "per-allele" – i.e., assuming a linear additive change across the three genotype categories. See DNA.
References
Other terms in 'Useful genetic terms ':
- Chromosome
- Cis- and trans-variants
- Copy number variation
- Deoxyribonucleic acid (DNA)
- Gene
- Genetic variant
- Genotype
- Haplotype
- Heterozygous or Heterozygote
- Homozygous or Homozygote
- Linkage disequilibrium (LD)
- Locus
- Palindromic single nucleotide polymorphism (SNP)
- Polygenic risk score (PRS)
- Polymorphism
- Rare variants
- Single nucleotide polymorphism (SNP)