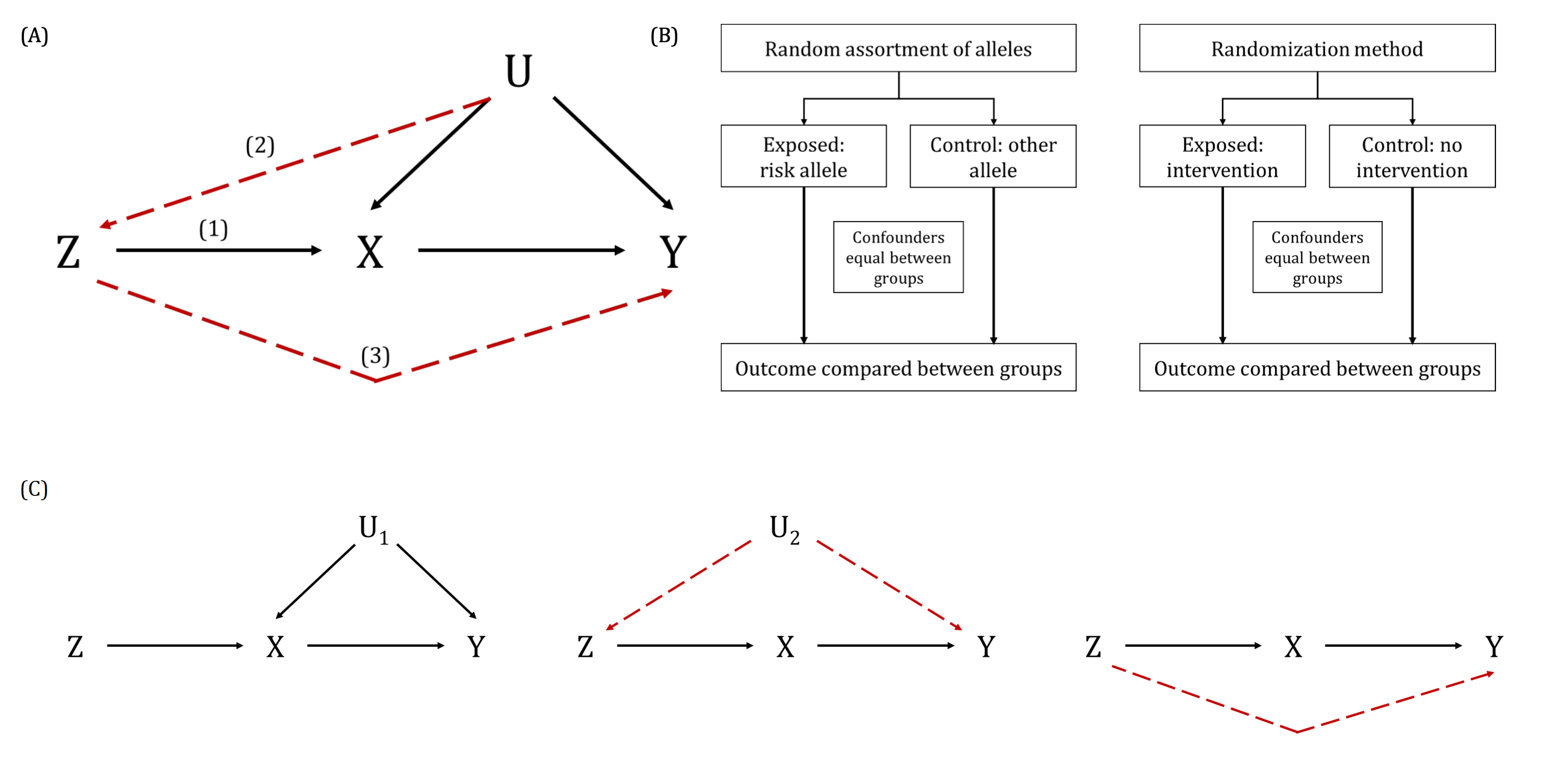
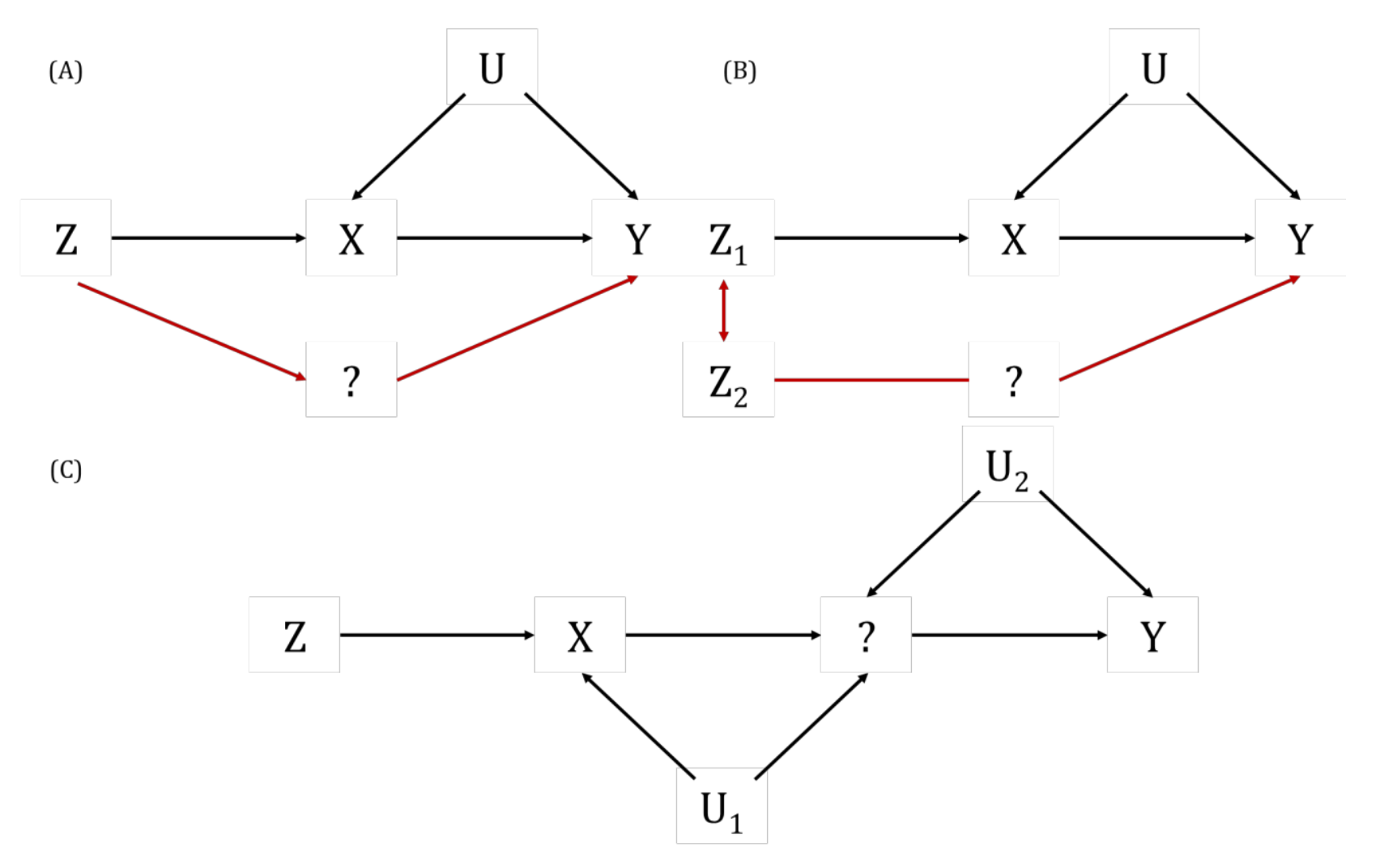
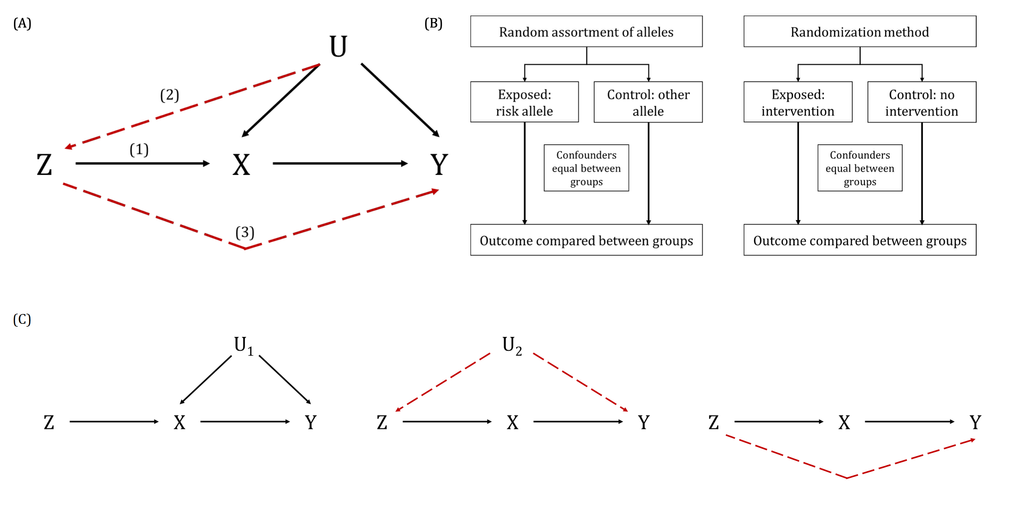
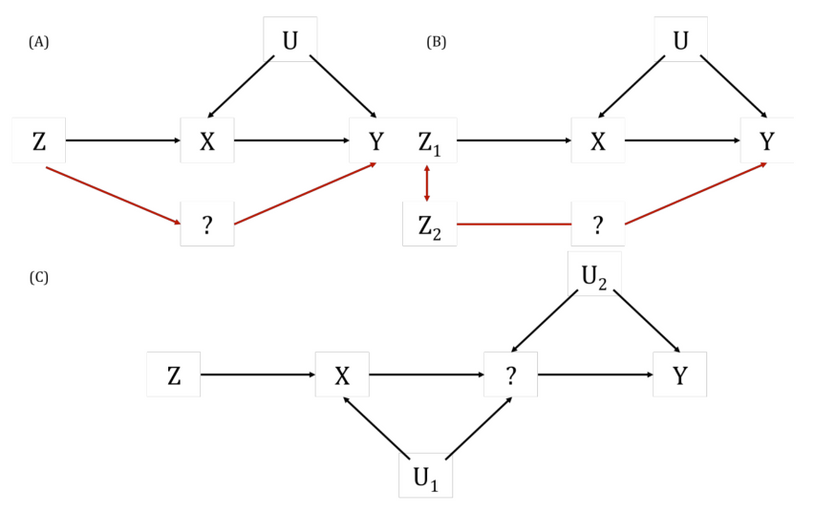
A key way in which the exclusion restriction assumption may be violated. Also known as “genuine” or “true” pleiotropy. In MR, this is when a genetic variant influences the outcome (directly or indirectly, through other traits) independently of the hypothesised exposure.
This can result in biased MR estimates because of violation of the exclusion restriction assumption. For example, if some or all of the genetic instrumental variables (IVs) that are robustly associated with an exposure of interest are also associated with other traits upstream of the outcome, independently of the exposure, then the MR estimate will be the combined (i.e., total) effect of all of these (independent) traits and not the (i.e., direct) effect of the exposure of interest alone. Methods such as MR-Egger, weighted median- and mode-based MR methods have been developed to explore and account for the impact of horizontal pleiotropy in MR studies. This can also be informative about the trait’s aetiology. There are further nuanced ways in which horizontal pleiotropy can lead to bias in MR analyses based on whether the pleiotropy is balanced or unbalanced, the latter of which is sometimes also known as directional pleiotropy. For example, if a subset of the genetic variants being used as instrumental variables (IVs) in an MR analysis have heterogenous causal effect estimates but these are equally balanced around the true causal effect (i.e., some have negative, some have positive effects), this balanced pleiotropy will likely not bias the resulting causal effect estimate. If, however, those genetic IVs have a consistent pleiotropic effect away from the true causal effect, this unbalanced pleiotropy will likely bias the derived causal effect estimate in a directional way.
References
- Davey Smith G, Hemani G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum Mol Genet 2014; 23: R89-R98.
- Zheng J, Baird D, Borges MC, et al. Recent Developments in Mendelian Randomization Studies. Curr Epidemiol Rep 2017; 4: 330-345.
Other terms in 'Sources of bias and limitations in MR':
- Assortative mating
- Canalization
- Collider
- Collider bias
- Conditional F-statistic for multiple exposures
- Confounding
- Exclusion restriction assumption
- F-statistic
- Harmonization (in two-sample MR)
- Homogeneity Assumption
- Independence assumption
- INstrument Strength Independent of Direct Effect (InSIDE) assumption
- Intergenerational (or dynastic) effects
- Monotonicity assumption
- MR for testing critical or sensitive periods
- MR for testing developmental origins
- No effect modification assumption
- NO Measurement Error (NOME) assumption
- Non-linear MR
- Non-overlapping samples (in two-sample MR)
- Overfitting
- Pleiotropy
- Population stratification
- R-squared
- Regression dilution bias (attenuation by errors)
- Relevance assumption
- Reverse causality
- Same underlying population (in two-sample MR)
- Statistical power and efficiency
- Vertical pleiotropy
- Weak instrument bias
- Winner's curse